Understanding the evolutionary processes of pathogenic mycobacteria to prevent the emergence of high health hazard variants
Crédits : A. Le Meur
Thesis defence
25/11/2025
09:30:00
Adrien Le Meur, ESE
IDEEV - Salle Rosalind Franklin
Thesis directed by Guislaine Refrégier (ESE, équipe Génétique et Ecologie Evolutives .
Mycobacterium tuberculosis is a pathogenic bacteria responsible for the tuberculosis disease. It is the leading cause of mortality by infectious disease. Tuberculosis treatment is a multi-drug therapy that must be taken daily for several months, which limits its effectiveness in developing countries. Furthermore, the bacteria is able to acquire additional resistances to antimycobacterial drugs. In order to keep track of the disease and identify antibiotic resistances, M. tuberculosis is extensively sequenced. More than 200,000 strains sequencing archives are publicly available. During my thesis, I developed Genotube, a bioinformatics pipeline designed to identify variants, predict antibiotic resistance and determine the lineage of M. tuberculosis strains from sequencing archives.
Using this pipeline, we genotyped more than 50,000 sequencing archives publicly available. We found an association between mutations in genes involved in replication, reparation and recombination and antibiotic resistance. These mutations were associated with a higher mutation rate and a higher frequency of antibiotic resistance acquisition in the close species M. smegmatis. The presence of these mutator phenotypes could play a role in the apparition of multi-drug resistance in M. tuberculosis.
We also used Genotube on smaller datasets. We confirmed the presence of a new lineage of M. tuberculosis in Central Africa. We also analyzed samples from children with tuberculosis in Ile-De-France. We identified recent transmis- sion events between children originating from the same countries or treated in the same centers.
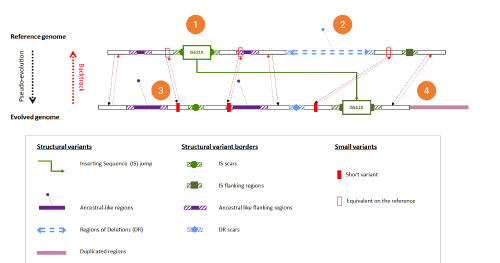
Lastly we studied the biases and artifacts introduced by the analysis of sequencing data. Standard benchmarks do not take into account the structural variant diversity of M. tuberculosis. We designed maketube, a bioinformatics tool that creates more realistic test genomes from what we know of M. tuberculosis diversity. Maketube creates large insertions, deletions, transposing sequence jumps and duplication regions. We show that maketube genomes diversity is much closer to the diversity found in natural strains compared to genomes created by standard benchmark methods. Using Make- tube, we tested several pipelines used for the genomics of M. tuberculosis and confirm a high precision but a lower recall. We show that insertions and duplications are an important source of false negatives, but not deletions. These re- sults will help researchers to choose between different bioinformatics protocols and filter out more effectively artefactual variants from their analyses.
Composition of the jury
- Maria Laura BOSCHIROLI, ANSES research director, reviewer and examiner
- Olivier TENAILLON, INSERM research director, reviewer and examiner
- Thibaut MOREL-JOURNEL, associate professor, examiner
- Gaelle LELANDAIS, associate professor, examiner
- Guislaine REFREGIER, associate professor, thesis director